Conducted as Genome Science Course before 2021
Fall 2022November 28 - 30, 2022
Fall 2021December 06-09, 2021
Fall 2020November 02-06 & 10, 2020
Canceled due to the Covid-19 outbreak.
Spring 2019June 03 - 07 & 11, 2019
Fall 2019 November 25-29 & December 02, 2019
2018 FallNovember 12-16, 2018
2018 SpringMay 28 - 31, 2018
Experiments and analyses: May 28 - 31
Preparation for presentation: June 01
Presentation of the results of the courses at international symposium: June 05
2017 FallNovember 13-17, 2017
2017 SpringMay 22-26, 2017 & May 31, 2017
2016 FallOctober 24-28, 2016






Based on the fecal DNA from Japanese macaques in Yakushima, students analyzed DNA (for sex identification) and steroid hormone (for evaluation of the stress level). They compared these laboratory results with parasite infection data collected in Yakushima.
Unfortunately, they couldn't find obvious correlation between parasite infection and stress hormone levels, but could learn how to analyze parasite, DNA and hormone from identical feces and enjoy combining field and laboratory works.
Takashi Hayakawa
2016 SpringMay 30 - June 3 & June 7, 2016


2015 Fall2015/10/26-2015/10/30, 11/05





A) Deer group
DNA and hormonal analyses of deer feces for the determination of sex and reproductive status By applying two molecular methods (DNA and hormonal analyses), participants will determine the sex and reproductive status of Yakushima deer from feces collected during the Field Science Course. First, deer genomic DNA will be extracted from the feces. The participants will determine the deer sex by amplification of genes located on the sex chromosomes. Second, participants will also analyze the sex steroid hormones (estrogen, progesterone, and testosterone) from the same deer feces to determine sex. In addition, by measuring the concentration of each hormone, the participants will estimate the reproductive status of deer. Because fall is the breeding season for deer, if female and male adults are in estrus and in rut, respectively, female (estrogen/progesterone) and male (testosterone) hormones will show high concentrations. Based on the results from these two molecular analyses, we will try to develop practical methods for estimating reproductive status of wild deer.
B) Plant group
Species composition and phenology in fern gametophyte For the gametophyte samples collected from Yakushima, we will try to identify the species or the genera to which they belong using molecular analysis. We will extract DNA from the samples and determine rbcL gene sequences from them. Subsequently, we will compare these sequences with registered DNA sequences in the gene bank.
2015 Spring2015/06/02-2015/06/07, 06/09



Following the Field Science Course will be the Advanced Laboratory Skills in Field Biology. If you are planning to take both the Field Science and Advanced Laboratory Skills in Field Biologys, we will be using samples collected from the Field Science Course in Yakushima Island for the Advanced Laboratory Skills in Field Biology. Therefore, we recommend that you take a Advanced Laboratory Skills in Field Biology accordingly to the Field Science course you took (if you take the Field Science Course). In addition, if you do not have experience with genome experiments, we recommend Advanced Laboratory Skills in Field Biologys A) - C). These combinations enable you to analyze the samples collected by yourselves. If you are interested in NGS and data analysis for NGS, take course D), regardless of the group you took in the Field Science Course (if you did take any Field Science Courses).
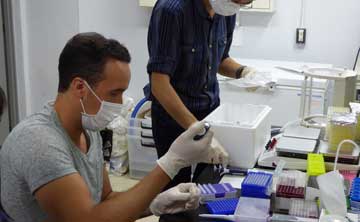
A) Individual identification of fecal DNA
We will focus on DNA individual identification method based on genotyping. DNA of monkeys and deer will be extracted from the feces collected during the Field Science Course. After DNA extraction, we will identify the sex using genetic marker and will identify individuals using several microsatellite loci.
B) Insect DNA barcoding
The Insect group will collect and identify insect species in Yakushima. Using these collected insects, we will sequence the mitochondrial DNA in order to provide a DNA barcode catalogue of insects in Yakushima. Based on the DNA barcode catalogue thus obtained, we will analyze the food repertories of Yakushima monkeys using the monkey feces-derived sequence data which were obtained by the members of 2014 Genome Training Course using MiSeq next generation sequencer.
C) Mushroom DNA barcoding
The Mushroom DNA group will collect and identify mushroom species in Yakushima. Using these collected mushrooms, we will sequence the ITS region of the genomic DNA in order to provide a DNA barcode catalogue of mushrooms in Yakushima Island.
D) Whole genome analyses of the Yakushima monkey
Lecturers will provide a whole genome shotgun (WGS) sequence of a Yakushima monkey, Macaca fuscata yakui determined by HiSeq2000 high-throughput sequencer. Using this data, we will try to analyze the functional gene sequences (e.g., sensory receptor gene repertoire), genetic diversity in the genome-wide scale, genetic differences with other macaque species, and historical changes of the effective population sizes of Yakushima monkeys in order to reveal the ecology and evolution of Yakushima monkeys. This group will not do "wet" laboratory experiments, but will focus on "dry" bioinformatics using personal- and super- computers.
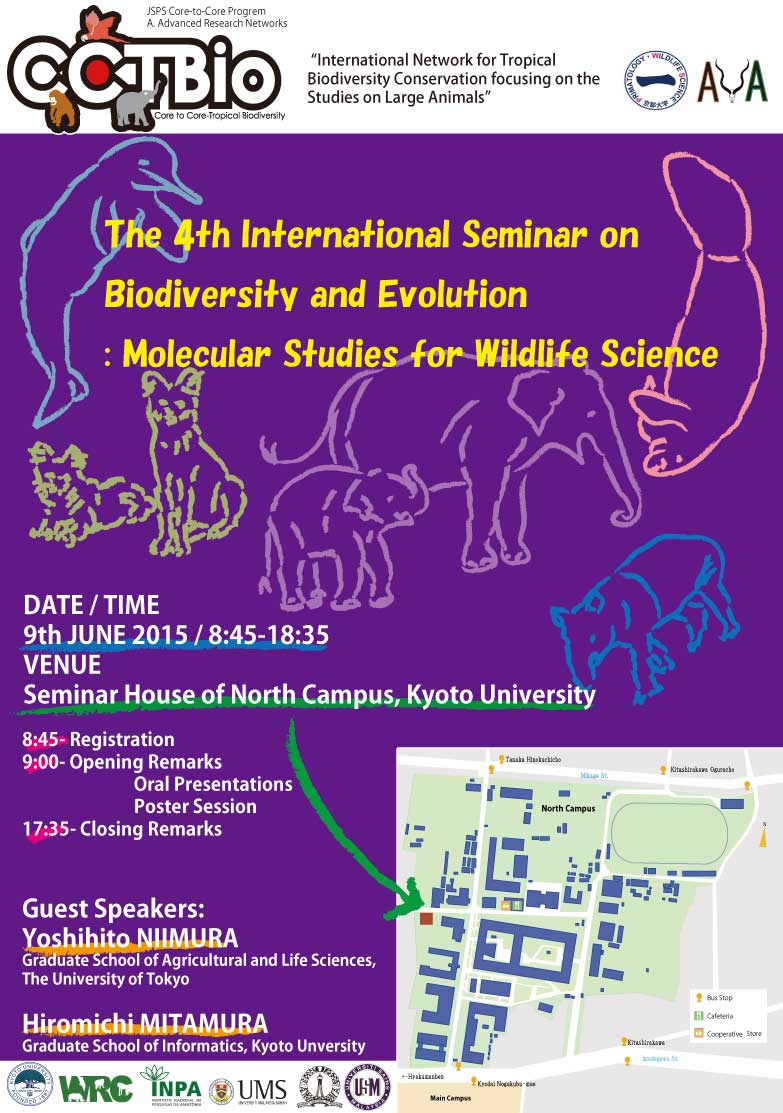
International Seminar
Date: June 9 (Tue), 2015
Place: Science Seminar House, Yoshida Campus of Kyoto University.
We will have talks from visiting foreign students and host researchers. We will also present the results of Field Science Course and Advanced Laboratory Skills in Field Biology in the poster session.

Introduction.pdf

Application Form.doc
Please read Introduction.pdf, and submit Application Form by e-mail during the following period.
Mail to: Ms. Sakai, sakai.yoko.2e
Period: April 1 to April 16, 2015 (deadline is April 16)
Guidance for the Field Science Course and Advanced Laboratory Skills in Field Biology, at WRC in Yoshida Campus. We will have a welcome party for participants from abroad.
2014 Fall2014/10/27 - 2014/10/31, 11/06
October 9: 13:00- Guidance for the course at PRI seminar room
October 27-31: Practice course at PRI
November 6: Presentation at PRI
Application Period: July 11 to July 31, 2014.
2014 Spring2014/05/29-2014/06/06 (excluding 2014/05/31 - 2014/06/01)



Presentation at the international seminar required at June 6.
Target re-sequencing of genome of Japanese monkeys from their feces, collected in Yakushima Island. Survey of micro-biota in the intestine of Japanese monkeys by next-generation sequencing. Identify insects species predated by monkeys by next-generation sequencing of their feces. Determine DNA sequence of insects collected in Yakushima Island for DNA bar-coding. Determine DNA sequence of ferns collected in Yakushima Island for DNA bar-coding.
Place: Science Seminar House, Kyoto University.
We will have talks of visiting foreign students and host researchers. We will also present the results of Field Science Course and Advanced Laboratory Skills in Field Biology.